Abstract
Introduction: Diffuse large B-cell lymphoma (DLBCL) is an aggressive B-cell neoplasm and the most common lymphoid malignancy worldwide. DLBCL, NOS is a genetically and biologically complex entity that remains uniformly treated at diagnosis despite the growing appreciation for subgroups with molecular distinctions. There are various approaches for assigning DLBCL into one or more molecular subgroups using either gene expression and/or mutation patterns. Several genetic classification systems have been proposed, each sharing commonalities. Distinctions between the definitions of individual classes and the ability to reproducibly assign individual patient samples to subgroups using each system remains a significant barrier to their implementation, further emphasized by up to 50% of patients that remain unclassified. The disparities among the genetic features considered by each classification scheme further limits the ability to consolidate, let alone adopt, these for research and clinical studies. We aimed to develop a classification system that unifies and extends the robustness of existing classification algorithms.
Methods: We assembled whole genome sequencing (WGS) data from 900 tumors from DLBCL, Burkitt (BL), follicular (FL) and other mature B-cell neoplasms to comprehensively determine the set of significantly mutated genes (SMG) and common targets of aberrant somatic hypermutation (aSHM). These were intersected with the features used by the current DLBCL classification systems and a curated set of lymphoma genes to tabulate the mutation matrix. Common structural rearrangements involving oncogenes were determined from WGS data and the copy number state of focal high-level recurrent alterations was united with simple somatic mutations. For genes containing mutation hot-spots each was considered separately.
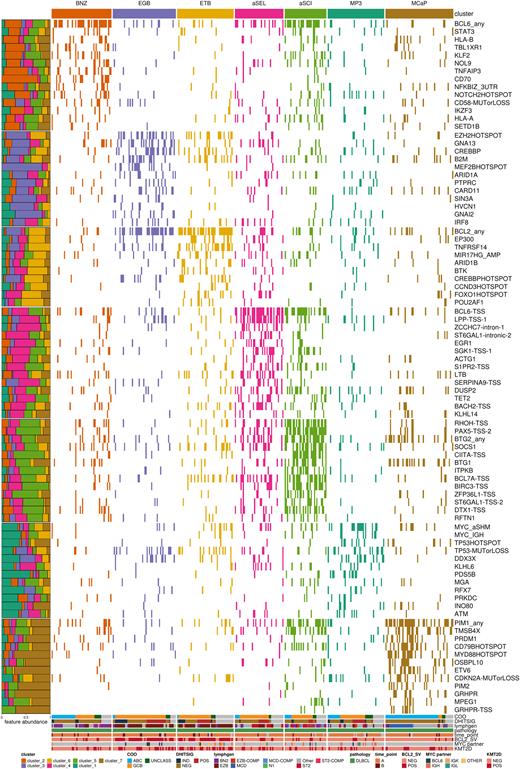
Results: Clustering all cases using non-negative matrix factorization (NMF) analysis resulted in a solution with 7 distinct genetic subgroups (Figure 1). Many of these directly resemble clusters in the existing systems with either expanded coverage or refinement of a subgroup into two. First, the BNZ cluster was highly enriched for any aberration in BCL6, 3'UTR mutations in NFKBIZ, and truncating mutations in NOTCH2. Unlike the similar BN2/C1/NOTCH2 groups defined previously, the NFKBIZ mutations were weighted more highly than those of NOTCH2 in defining the BNZ subgroup. The cluster we named MCaP resembled the MCD/C5/MYD88 group. Tumors classified in this subgroup had evidence of aSHM with frequent non-coding mutations in PIM1,CD44, GRHPR, PIM2, and BCL11A among others. Another cluster, MP3, was enriched for coding mutations or aSHM in the MYC TSS (MYC_aSHM), which were generally correspondent with IG::MYC rearrangements. MP3 tumors were characterized by TP53 mutations, including hotspots, and significantly depleted for aberrations in BCL2, BCL6 and lower mutation rates at aSHM targets SGK1, PAX5, CIITA, RHOH and others. Notably, tumors in this subgroup were enriched for IRF4 translocations, suggesting potential overlap with large B-cell lymphoma with IRF4 rearrangement.
Our approach further refined both the the ST2/C4 and EZB/C3/BCL2 classes into two smaller subgroups, aSEL/aSCI and EGB/ETB, respectively. The EGB tumors were enriched for GNAI2 mutations, and MYC translocations involving the BCL6 enhancer, and were more often associated with MEF2B hotspot mutations. Notably, ETB was significantly enriched for mutations in the lysine acetyltransferase (KAT) domain in CREBBP, while EGB was significantly enriched for non-KAT domain mutations. ETB tumors were further characterized by mutations in STAT6 and FOXO1 hotspot regions, MYC rearrangements with other non-IG partners, and TNFRSF14 mutations, largely resembling the mutational profile of follicular lymphomas.
Conclusions: We present here a refined classification of DLBCL genetic subgroups that combines previously described classification algorithms and extends it to allow classification of more tumors into additional subgroups that preserve the major divisions of existing systems. This represents an important advancement that will facilitate further understanding of genomic complexity in this disease.
Disclosures
Coyle:Dren Bio, Inc.: Consultancy; Allakos, Inc.: Consultancy. Rushton:SAGA Diagnostics: Consultancy. Scott:Abbvie: Consultancy; Janssen: Consultancy, Research Funding; NanoString: Patents & Royalties; AstraZeneca: Consultancy, Honoraria; Incyte: Consultancy; Roche: Research Funding. Steidl:Abbvie: Consultancy; Bayer: Consultancy; Bristol Myers Squibb: Consultancy; Curis Inc: Consultancy; Epizyme: Research Funding; Roche: Consultancy; Seattle Genetics: Consultancy; Trillium Therapeutics: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal